Triangle
MRSEC investigators Zauscher,
Chilkoti , and Yingling
recently developed a new, bioinspired method
to synthesize high molecular weight, single-stranded DNA (ssDNA).[1] This
enzyme-catalyzed
polymerization of ssDNA is conceptually similar to controlled
polymerization
of synthetic polymers, in which monomers are added to a growing chain one at a
time, and yields polynucleotides with narrow molecular
weight distributions (Fig. 1).
They now seek to leverage this method and develop,
aided by coarse-grained, dissipative particle dynamics (DPD) simulations,
a
library of copolymer architectures that can self assemble into a broad range of
morphologies. The
ability to synthesize polynucleotides that form complex nano- to meso-scale
morphologies,
including segregated, incompatible subdomains, or that directly contain nucleic
acid based drugs, has great potential for bionanotechnology and for
drug delivery
applications.
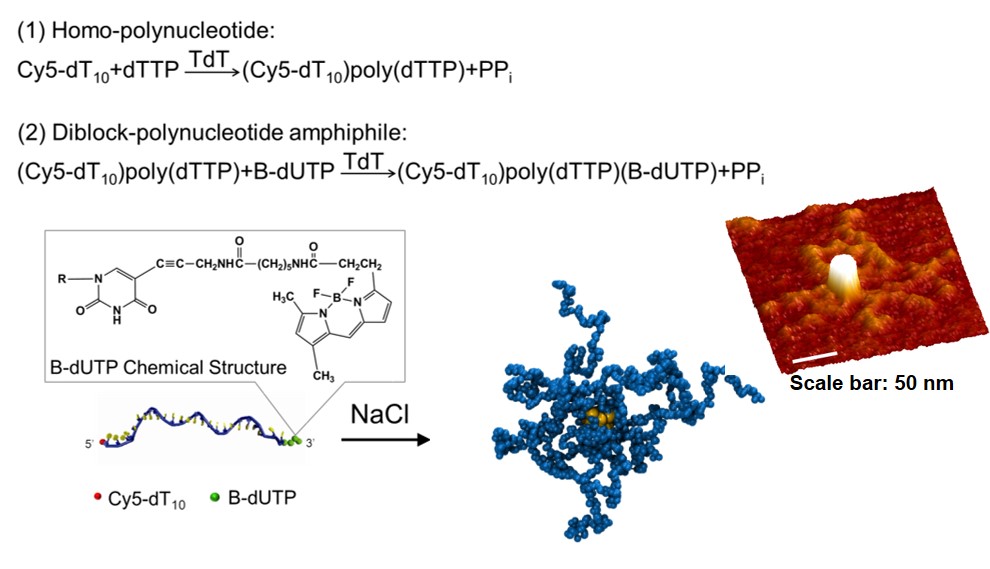
Figure
1:
Two-step, enzymatic polymerization of amphiphilic
polynucleotides using a template independent TdT polymerase.
Self-assembly into star-like micelles
shown by DPD simulation and high resolution AFM imaging.