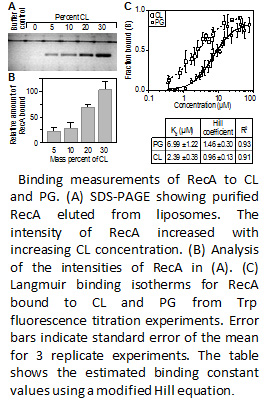
 Researchers in IRG3 of the Wisconsin MRSEC are studying bacteria to understand principles by which nature designs materials to achieve dynamic spatial targeting of molecules. Recently, the researchers have discovered that the spatial organization of phospholipids at the curved regions of bacterial cytoplasmic membranes has an important physiological role in bacteria, including the regulation and positioning of the universal enzyme RecA. RecA is a widely conserved cytoplasmic protein that performs a range of functions in Escherichia coli, including: DNA repair, homologous recombination, and induction of the SOS pathway. The IRG tested the hypothesis that the localization and function of the large, polar fraction of RecA in E. coli depends on its interaction with anionic phospholipids that accumulate at the large, negatively curved region of the cytoplasmic membrane. Figure 1 shows that RecA binds to the anionic phospholipids phosphatidylglycerol (PG) and cardiolipin (CL) with an apparent Kd of 7.0 and 2.4 uM, respectively. In addition it was determined that CL negatively regulated the strand exchange activity of RecA by inhibiting its ability to hydrolyze ATP in vitro. Intriguingly, the inhibition of ATP hydrolysis by RecA was dependent on its C-terminal auto-regulatory domain. CL-deficient strains of E. coli displayed elevated rates of accumulating spontaneous mutations compared to wild-type E. coli, which is consistent with the unregulated activation of RecA. These studies provide the IRG with insight into how membrane strain is connected to the organization of phospholipids and the biological function of the resulting material. The extension of these concepts to designing functional strained LC materials is underway.
Researchers in IRG3 of the Wisconsin MRSEC are studying bacteria to understand principles by which nature designs materials to achieve dynamic spatial targeting of molecules. Recently, the researchers have discovered that the spatial organization of phospholipids at the curved regions of bacterial cytoplasmic membranes has an important physiological role in bacteria, including the regulation and positioning of the universal enzyme RecA. RecA is a widely conserved cytoplasmic protein that performs a range of functions in Escherichia coli, including: DNA repair, homologous recombination, and induction of the SOS pathway. The IRG tested the hypothesis that the localization and function of the large, polar fraction of RecA in E. coli depends on its interaction with anionic phospholipids that accumulate at the large, negatively curved region of the cytoplasmic membrane. Figure 1 shows that RecA binds to the anionic phospholipids phosphatidylglycerol (PG) and cardiolipin (CL) with an apparent Kd of 7.0 and 2.4 uM, respectively. In addition it was determined that CL negatively regulated the strand exchange activity of RecA by inhibiting its ability to hydrolyze ATP in vitro. Intriguingly, the inhibition of ATP hydrolysis by RecA was dependent on its C-terminal auto-regulatory domain. CL-deficient strains of E. coli displayed elevated rates of accumulating spontaneous mutations compared to wild-type E. coli, which is consistent with the unregulated activation of RecA. These studies provide the IRG with insight into how membrane strain is connected to the organization of phospholipids and the biological function of the resulting material. The extension of these concepts to designing functional strained LC materials is underway.