 In designing new motile materials, much can be learned by studying the physical mechanisms underlying cell crawling. One important form of cell crawling is driven by self-assembly of the protein actin. In this process, energy is supplied and various proteins cooperate to assemble actin from small proteins into a branched network. We have conducted the first physically-consistent simulations of this process and have discovered that the mechanism driving motion of the cell boundary (modeled in our case by a flat disk) is very simple: the buildup of the branched actin network behind the disk drives the disk forwards because the disk is repelled by actin. This is reminiscent of the old joke about why bagpipe players always walk while they play (to get away from the noise). Understanding of this mechanism opens the way to the development of new materials that can move, such as asymmetrically-coated beads in self-generated concentration gradients.
In designing new motile materials, much can be learned by studying the physical mechanisms underlying cell crawling. One important form of cell crawling is driven by self-assembly of the protein actin. In this process, energy is supplied and various proteins cooperate to assemble actin from small proteins into a branched network. We have conducted the first physically-consistent simulations of this process and have discovered that the mechanism driving motion of the cell boundary (modeled in our case by a flat disk) is very simple: the buildup of the branched actin network behind the disk drives the disk forwards because the disk is repelled by actin. This is reminiscent of the old joke about why bagpipe players always walk while they play (to get away from the noise). Understanding of this mechanism opens the way to the development of new materials that can move, such as asymmetrically-coated beads in self-generated concentration gradients.
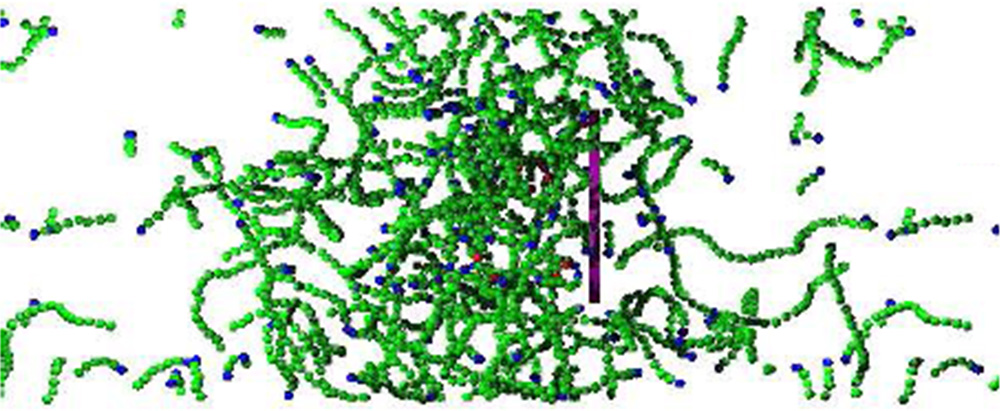
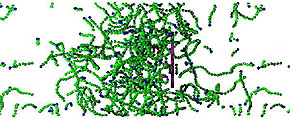
Figure: Snapshot from simulation of the disk (purple) moving to the right due to assembly of a branched actin network to its left. The green spheres represent actin proteins in filaments.